Blog Categories
Decoding Arterial and Venous Vascular Biology with BxGenomics Bulk RNA-Seq View
| Understanding the molecular differences between arterial and venous blood vessels is essential for advancing vascular biology and developing targeted therapies for cardiovascular disease. In a recent International Journal of Molecular Sciences study, researchers from BioInfoRx and the University of North Carolina at Chapel Hill used the BxGenomics' Bulk RNA-Seq View to perform a comprehensive bulk RNA-seq analysis of porcine arterial and venous vascular smooth muscle cells (ApSMCs and VpSMCs), uncovering hundreds of differentially expressed genes (DEGs) and pathway-level differences that help explain vessel-specific behaviors. |  |
SpaceSequest—Bridging Spatial Transcriptomics Platforms with a Single Comprehensive Pipeline
| Spatial transcriptomics has rapidly emerged as one of the most transformative innovations in modern biology, empowering researchers to map gene expression while preserving the intricate architecture of tissues. Recent advances in the field have resulted in a variety of new platforms including Visium, Visium HD, and Xenium from 10x Genomics, and GeoMx and CosMx from NanoString Technologies (now part of Bruker). Developed by BioInfoRx in collaboration with Biogen, SpaceSequest emerges as a unified and elegant solution designed to harmonize data from all five major spatial transcriptomics platforms. |  |
Rebuilding the Airway Cell Atlas from a Landmark Nature Study Using Cellxgene VIP in BxGenomics
| In the high-impact Nature study “Local and systemic responses to SARS-CoV-2 infection in children and adults (Yoshida et al.,2021)”, scientists uncovered why children often fare better than adults when infected with COVID-19. At Cellxgene VIP (Li et al., 2022), we brought this discovery to life—moving beyond reading the findings to reconstructing them through dynamic, interactive visualizations. Cellxgene VIP is an interactive frontend visualization plugin for the cellxgene framework allowing researchers to generate a comprehensive set of high resolution plots with fully customizable settings in real time. It is part of BxGenomics, an integrated online platform developed by researchers at BioInfoRx for the analysis and visualization of both single-cell and bulk RNA-seq data. | 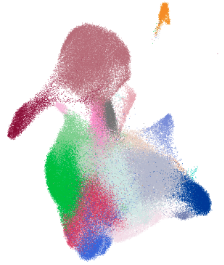 |
Expanding Horizons: BxGenomics Adds NanoString CosMx Data Analysis to scRNA-Seq View
| In a significant advancement for single-cell and spatial transcriptomics, BxGenomics, an integrated online genomics data analysis system developed by BioInfoRx, has expanded its capabilities by incorporating NanoString CosMx Spatial Molecular Imaging (SMI) data analysis into its scRNA-Seq View application. This integration marks a pivotal step toward providing researchers with a more comprehensive and streamlined web-based platform for analyzing high-dimensional single-cell spatial transcriptomics data. | 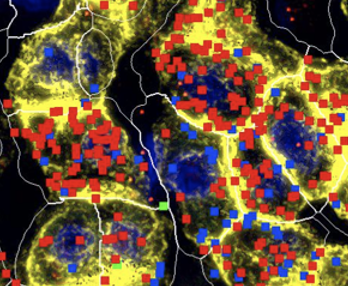 |
ScaleSC: Supercharging Single Cell RNA-seq Analysis with GPU Power
| Single cell RNA sequencing(scRNA-seq) has transformed our understanding of cellular biology by allowing us to examine gene expression in individual cells. However, this powerful technology comes with a significant computational cost. In a revolutionary development for the single cell genomics community, Zhengyu Ouyang at BioInfoRx, along with researchers from Biogen, has introduced ScaleSC, a GPU-accelerated solution for single cell data processing (Hu et al., 2025). ScaleSC is freely available on GitHub, BioInfoRx also plans to integrate ScaleSC into the BxGenomics single-cell workflow. | 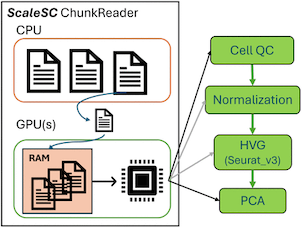 |
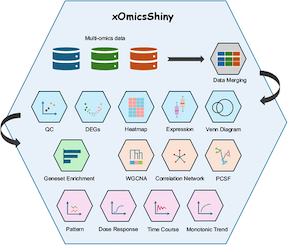
Unlocking the Power of Multi-Omics Data with xOmicsShiny
|
In the age of high-throughput biological research, multi-omics integration is essential for unraveling complex biological processes. However, managing, analyzing, and extracting meaningful insights from vast and complex datasets remains a challenge. xOmicsShiny (Gao et al., 2025), developed by researchers from BioInfoRx and Biogen, provides a comprehensive platform for cross-omics data analysis. Unlike existing tools, xOmicsShiny offers an intuitive interface for seamless data merging and interactive visualization across transcriptomics, proteomics, metabolomics, and lipidomics datasets, allowing researchers to efficiently explore pathway-level insights. Users of BxGenomics may also enjoy some of the features from xOmicsShiny in the future. |
 |
BxGenomics Revolutionizes HIV Microglia Research
The persistent challenge of curing HIV lies in its ability to hide in latent reservoirs, especially in the central nervous system (CNS). In a groundbreaking study presented through a collaboration between Zhengyu Ouyang, director of Bioinformatics at BioInfoRx, Johannes Schlachetzki from UC San Diego (UCSD), and other scientists, BxGenomics played a pivotal role in unraveling the molecular dynamics of microglia in people with HIV under antiretroviral therapy (ART).
5 Tips for Robust and Reproducible scRNA-Seq Data Analysis
BioInfoRx and Biogen have developed scRNASequest, an open-source ecosystem for scRNA-seq analysis, visualization, and publishing. To make this powerful pipeline more accessible, our team has introduced a fully supported and user-friendly version in BxGenomics, enabling academic and small biotech researchers to leverage its capabilities without the need for extensive IT and bioinformatics expertise. Here we shared some tips we learned from developing and running the pipeline on hundreds of scRNA-Seq experiments.
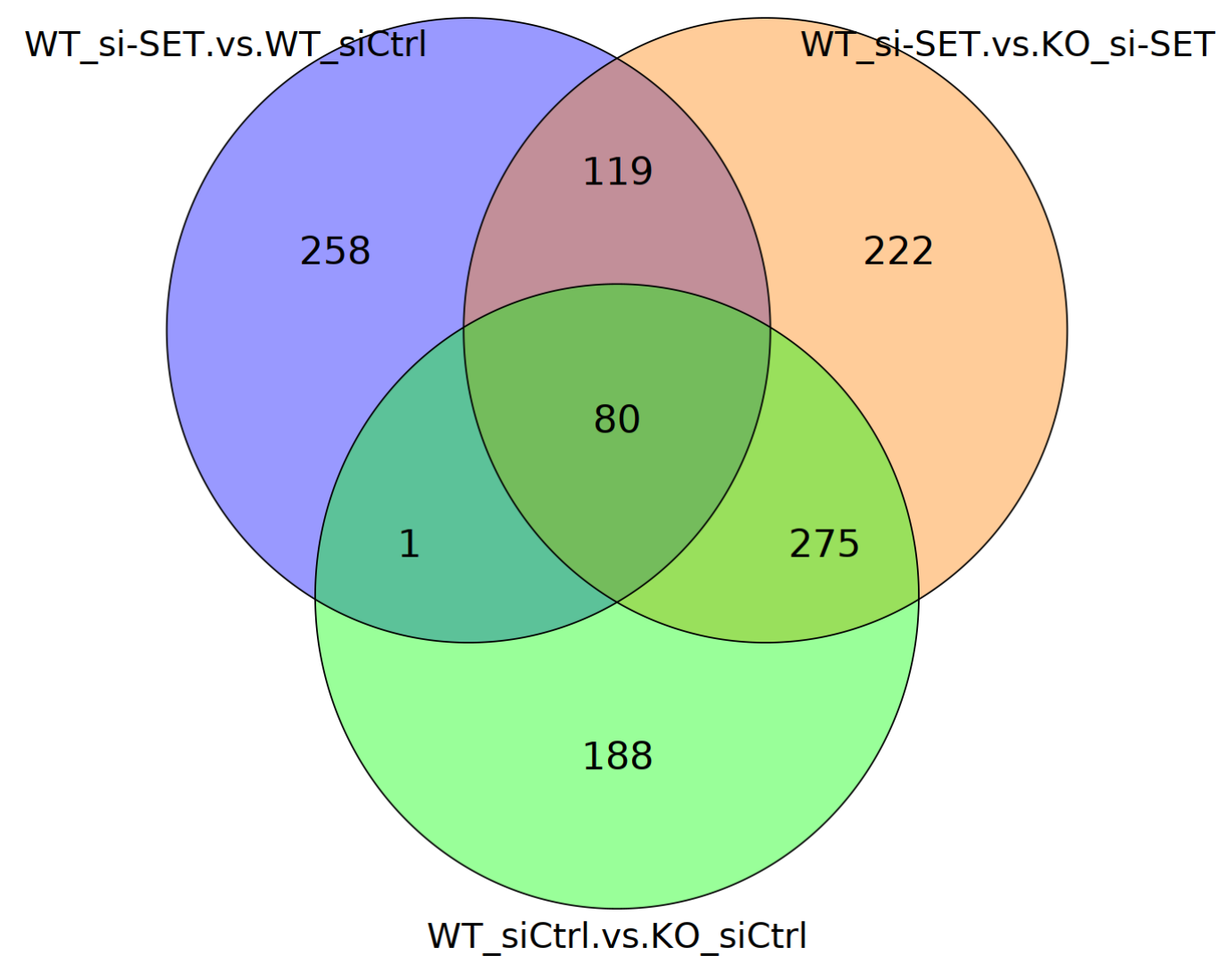
RNA-Seq Case Study: Genes Regulated by p53-SET Interplay
|
The oncoprotein SET can bind to p53 and inhibits p53 transcriptional activity in unstressed cells. To identify the genes regulated by SET in a p53-dependent manner, the endogenous SET was depleted by siRNA in control or p53 knockout U2OS cells. We use BxGenomics DEG and Venn Diagram tools to find genes regulated by p53-SET. Reference: Nature. 2016 Oct 6;538(7623):118-122. doi: 10.1038/nature19759. |
 |
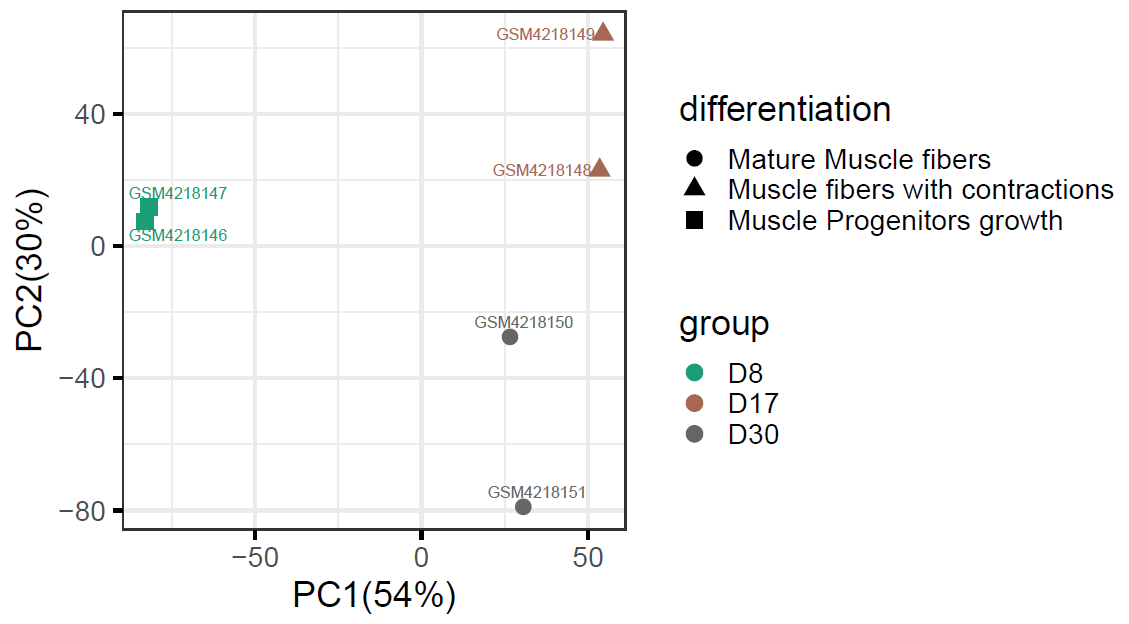
RNA-Seq Case Study: iPSC Differentiation into Muscle and Neuron
|
We use BxGenomics to examine RNA-Seq data from three key time points during human iPSC differentiation into mature muscle fibers with motor neurons. We can detect consistent genes changes as the cells differentiate, and myogenesis and nurogenesis genes are highly enriched in later time points. Reference: Cells. 2020 Jun 23;9(6):1531. doi: 10.3390/cells9061531. |
 |