Blog Categories
BxGenomics Revolutionizes HIV Microglia Research
BxGenomics is an integrated online system with scRNASequest (Li et al., 2023) and Cellxgene VIP (Li et al., 2022). scRNASequest is an end-to-end pipeline designed for single-cell RNA-seq data analysis, visualization, and publishing. Cellxgene VIP (Visualization In Plugin) is an interactive frontend visualization plugin for the cellxgene framework, significantly expanding its capabilities. It allows researchers to generate a comprehensive set of high resolution plots with highly customizable settings in real time, access advanced analytical functions, perform analysis in a Jupyter Notebook-like environment, if desired, and visualize multi-modal data with enhanced flexibility.
The persistent challenge of curing HIV lies in its ability to hide in latent reservoirs, especially in the central nervous system (CNS). In a groundbreaking study presented through a collaboration between Zhengyu Ouyang, director of Bioinformatics at BioInfoRx, Johannes Schlachetzki from UC San Diego (UCSD), and other scientists, BxGenomics played a pivotal role in unraveling the molecular dynamics of microglia in people with HIV under antiretroviral therapy (ART)(Schlachetzki et al., 2024).
In this study, high quality microglia were isolated from the dorsolateral prefrontal cortex (DLPFC) collected from three male participants (Fig. 1A) enrolled in UCSD’s unique “Last Gift” rapid autopsy program. The program closely followed people with HIV until days or even hours before death. These three people had chronic HIV and were on long-term ART (Fig. 1B). CD45+ cells were isolated (Fig. 1C), and multi-omics genomics libraries were performed.

Figiure 1 Experimental design and single-cell transcriptomics of CD45+ cells isolated from the cortex of individuals with HIV under antiretroviral therapy.
Single-cell RNA-seq data generated using the microfluidic-based 10X Chromium platform were analyzed through BioInfoRx's scRNASequest. Sequencing reads were processed with CellRanger and the resulting count matrices, labeled with gene symbols and cell barcodes, were imported into R using Seurat. After QC and removal of an outlier cluster, a total of 25,091 CD45+ cells were clustered into 15 clusters (Fig. 1D). Within Cellxgene VIP, all brain myeloid cells expressed to varying degrees CX3CR1, P2RY12, TREM2, GPR34, and CSF1R, as shown in Fig. 1E.
Thanks to Cellxgene VIP, feature plots highlight the expression of marker genes in individual clusters (Fig. 2A). IFI27, associated with antigen presentation and interferon signaling, is expressed in Cluster 3; the stress response gene JUN is expressed in Cluster 4; the pro-inflammatory gene IL1B is expressed in Cluster 6; and the chemokine and cytokine signaling gene CCL4 is expressed in Cluster 7. The interactive plotting function in Cellxgene VIP also helps researchers generate a dot plot depicting marker genes for each cluster (Fig. 2B). The size of the circles indicates the fraction of cells expressing the genes, while the color scale represents the mean expression level per group.
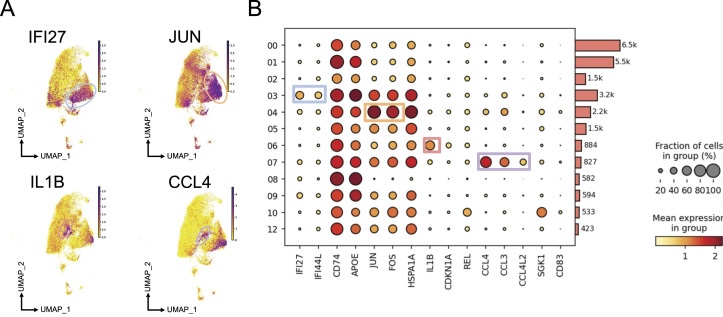
Figure 2. Characterization of microglial phenotypes in the cortical regions of individuals with HIV under antiretroviral therapy based on gene expression.
Besides single cell RNA-seq data, single cell ATAC-Seq data was also processed through our single cell ATAC-Seq pipeline which helps to identify active gene regulatory elements in HIV-infected microglia. The single cell chromatin accessibility map of isolated CD45+ cells were visualized and presented in Cellxgene VIP. DNA motifs enriched for PU.1/ETS family members, which are lineage-determining transcription factors of microglia, were successfully identified.
This research highlights the significant impact of the BxGenomics in advancing HIV cure strategies. Computational tools enable high-resolution, single-cell analysis of microglia, paving the way for targeted therapies that address persistent viral reservoirs in the brain. The research shows that microglial populations were heterogeneous regarding their gene expression profiles but showed similar chromatin accessibility landscapes. Strategies for HIV cure and addressing neurocognitive impairment must account for the myeloid population to be successful.
References:
Schlachetzki, J. C. M., Gianella, S., Ouyang, Z., Lana, A. J., Yang, X., O'Brien, S., Challacombe, J. F., Gaskill, P. J., Jordan-Sciutto, K. L., Chaillon, A., Moore, D., Achim, C. L., Ellis, R. J., Smith, D. M., & Glass, C. K. (2024). Gene expression and chromatin conformation of microglia in virally suppressed people with HIV. Life Science Alliance, 7(10), e202402736. https://doi.org/10.26508/lsa.202402736
Li, K., Sun, Y. H., Ouyang, Z., Negi, S., Gao, Z., Zhu, J., Wang, W., Chen, Y., Piya, S., Hu, W., Zavodszky, M. I., Yalamanchili, H., Cao, S., Gehrke, A., Sheehan, M., Huh, D., Casey, F., Zhang, X., & Zhang, B. (2023). scRNASequest: An ecosystem of scRNA-seq analysis, visualization, and publishing. BMC Genomics, 24, 228. https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-023-09332-2
Li, K., Ouyang, Z., Chen, Y., Gagnon, J., Lin, D., Mingueneau, M., Chen, W., Sexton, D., & Zhang, B. (2022). Cellxgene VIP unleashes full power of interactive visualization and integrative analysis of scRNA-seq, spatial transcriptomics, and multiome data. bioRxiv. https://doi.org/10.1101/2020.08.28.270652