Blog Categories
Rebuilding the Airway Cell Atlas from a Landmark Nature Study Using Cellxgene VIP in BxGenomics
In the high-impact Nature study “Local and systemic responses to SARS-CoV-2 infection in children and adults (Yoshida et al.,2021)”, scientists uncovered why children often fare better than adults when infected with COVID-19. At Cellxgene VIP (Li et al., 2022), we brought this discovery to life—moving beyond reading the findings to reconstructing them through dynamic, interactive visualizations.
Cellxgene VIP is an interactive frontend visualization plugin built on the cellxgene framework allowing researchers to generate a comprehensive set of high resolution plots with fully customizable settings in real time. It is part of BxGenomics, an integrated online platform developed by researchers at BioInfoRx for the analysis and visualization of both single-cell and bulk RNA-seq data. The study was built on a powerful dataset—over 650,000 single cells profiled from matched nasal, tracheal, bronchial, and blood samples—identifying 59 distinct cell types and states in the airways and 34 in the blood. In this blog, we focus specifically on the airway epithelial cells. The process of reconstructing some of the findings is recorded in a video and can be viewed here.
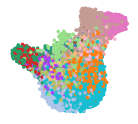
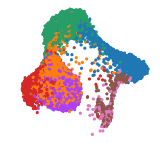
The epithelial cell populations are organized into two primary domains: one comprising ciliated cells and the other encompassing the basal-to-secretory cell differentiation pathway. Utilizing Uniform Manifold Approximation and Projection (UMAP), the study illustrates the extensive diversity of airway epithelial cell types—including multiple basal, goblet, ciliated, and transitional epithelial populations (Transit epi 1 and 2)—reflecting the inherent plasticity of the airway compartment. Through Cellxgene VIP, we reconstructed these UMAPs in detail (Fig. 1). Log in as a guest user or with a trial account to access the orginal data at: https://app.bxgenomics.com/bxg/app/scrnaview/app_project_launcher.php?ID=853. The platform enables users to explore the data interactively by hovering, zooming, and toggling between different age groups or infection states, offering insights beyond the capabilities of static journal figures.
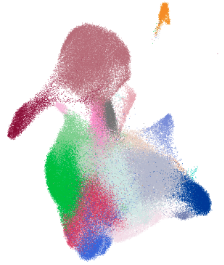 |
 |
 |
 |
 |
 |
 |
 |
Figure 1. Uniform Manifold Approximation and Projection (UMAP) visualization of annotated airway epithelial cells and immune cells
To validate and refine the cell type annotations used throughout the analysis, the authors presented dot plots of marker gene expression across annotated airway epithelial cell types, one of which was regenerated in Cellxgene VIP and is shown in Fig. 2. Each dot conveys two key metrics: its size reflects the fraction of cells expressing a given gene, while its color intensity indicates the average expression level. For example, basal1 cells are marked by strong expression of KRT15 and DLK2, while goblet cells are defined by MUC5AC, and club cells have high expression in SCGB1A1 and SCGB3A1. Ciliated cells express canonical markers such as PIFO, reflecting their role in mucociliary clearance. Importantly, the study also highlights transitional states, such as transit epithelial cells—which bridge secretory and ciliated lineages. These cells co-express ciliated cell markers (e.g., PIFO) and secretory markers (e.g., MUC5AC) and are especially enriched in infected samples, reflecting epithelial remodeling in response to viral injury.
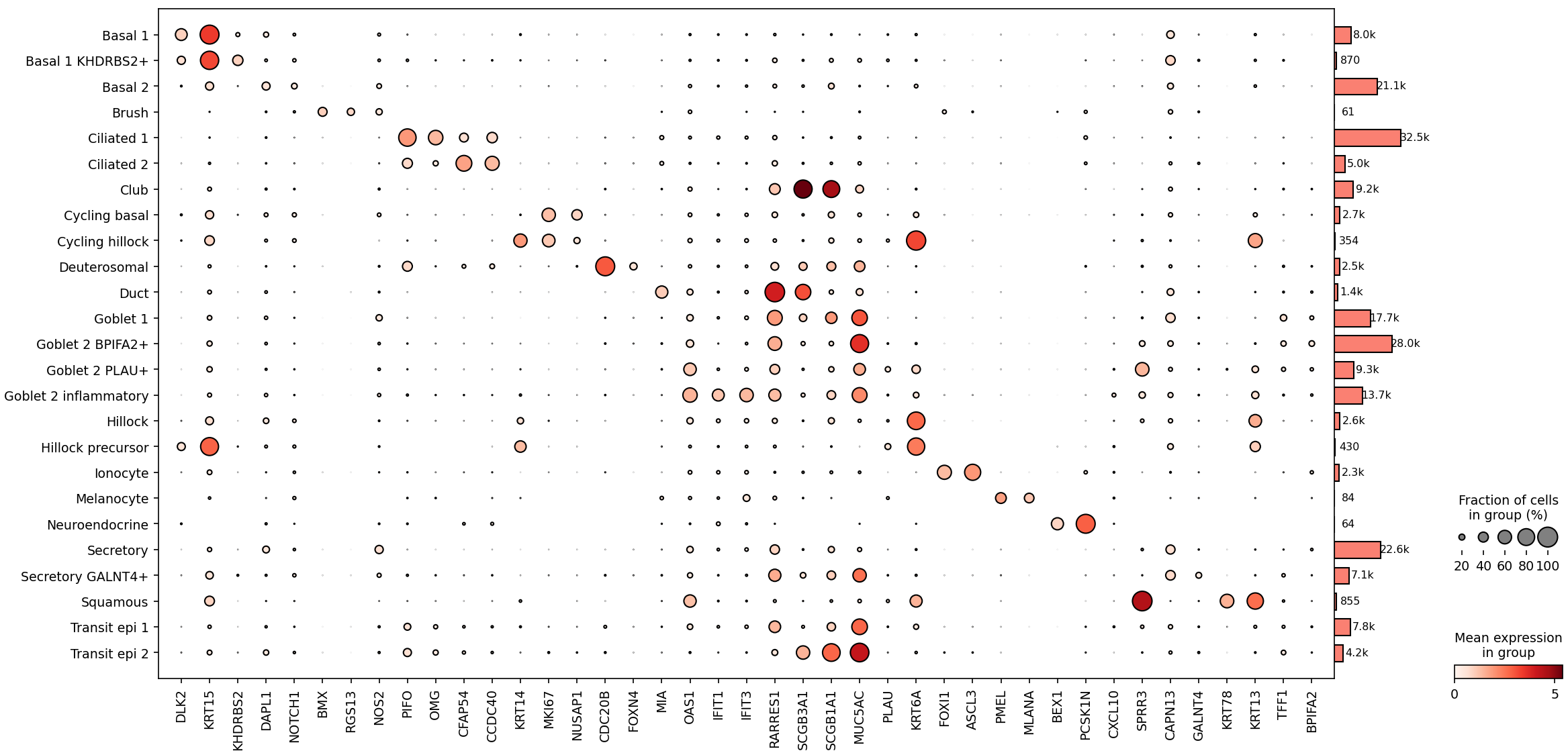
Figure 2. A dot plot showing marker genes for annotated airway epithelial cells
Beyond the transit epithelial cells, three additional populations were identified as differentiating toward ciliated cells: Deu (deuterosomal), Ba-d (basal differentiating) and IRC (interferon responsive cells). Marker genes for these populations, along with markers for transit epithelial cells (Transit epi 1 and 2) are shown in Fig. 3. CCNO and FOXN4 are marker genes for Deu, KRT4 and KRT7 are for Ba-d, IFIT1 and CXCL10 for IRC and S100A8 and s100A9 for transit epithelial cells. In the original paper, three marker genes are selected for each cell type; here, in Cellxgene VIP, only two markers are checked for each cell type.
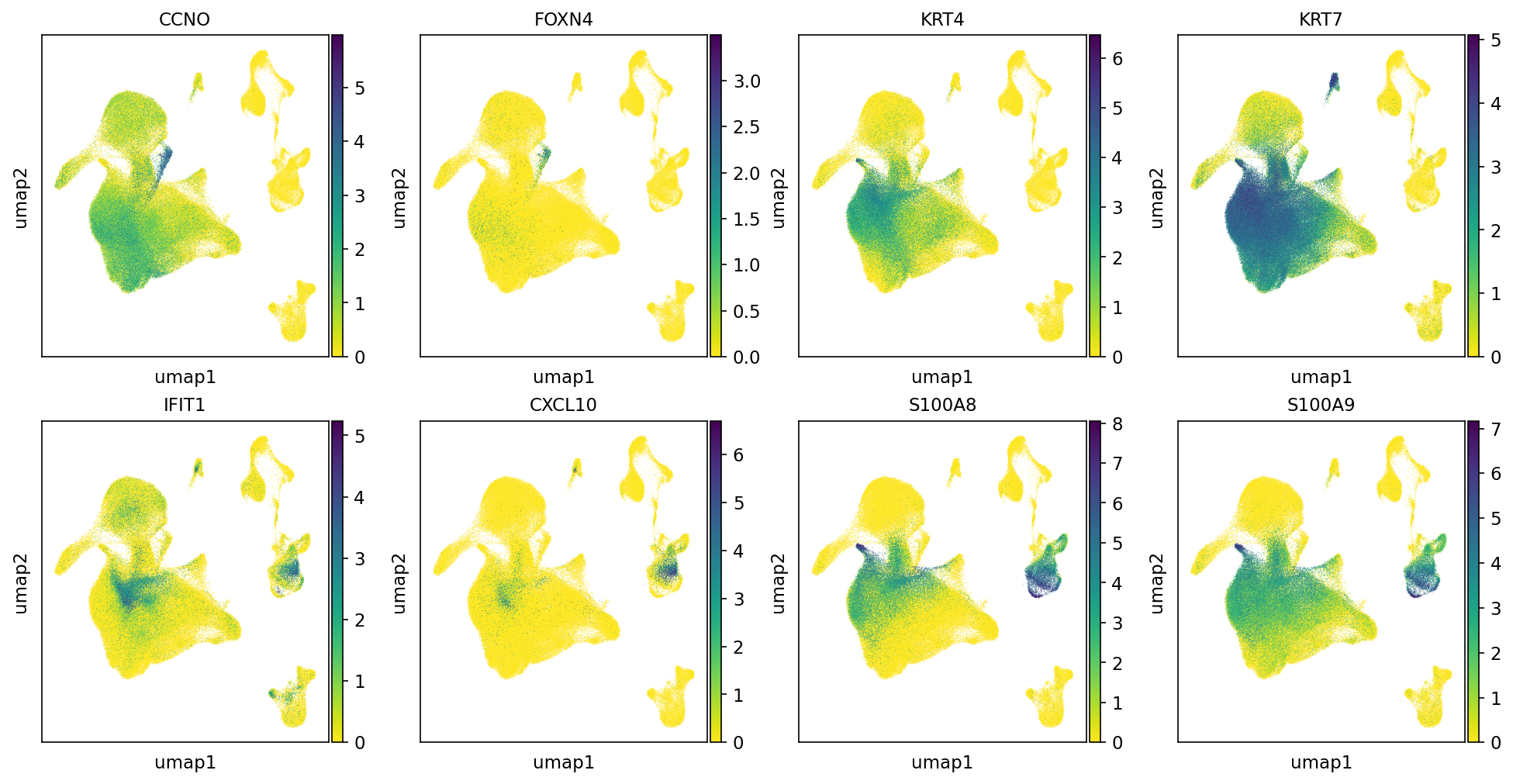
Figure 3. Marker genes for the three populations identified as differentiating to ciliated cells and markers of transit epithelial cells (Transit epi 1 and 2). Deu; deuterosomal, Ba-d; basal differentiating, IRC; interferon responsive cell.
With such finely resolved cell classifications, this analysis lays the foundation for deeper investigations—such as comparing cell type proportions between patients and healthy individuals, or across different age groups. Of particular interest are the transitional epithelial states, which may hold important clues to how the airway epithelium responds to and recovers from viral injury. While this blog focused on airway epithelial cells, the original study also identified 34 distinct cell types in blood, offering rich insights into systemic immune responses that are equally worth exploring. Due to space constraints, we have not reproduced all visualizations here—but the full dataset is available for detailed exploration. If you’re interested in diving deeper, we invite you to explore the complete dataset using Cellxgene VIP.
The tutorial for generating the three figures in Cellxgene VIP is available here: https://www.youtube.com/watch?v=_kiJ7rRbJC8
References:
Yoshida, M., Worlock, K.B., Huang, N., Lindeboom, R.G.H., Butler, C.R., Kumasaka, N., Dominguez Conde, C., Mamanova, L., Bolt, L., Richardson, L., et al. (2021). Local and systemic responses to SARS-CoV-2 infection in children and adults. Nature 597, 283–288. https://doi.org/10.1038/s41586-021-04345-x
Li, K., Ouyang, Z., Chen, Y., Gagnon, J., Lin, D., Mingueneau, M., Chen, W., Sexton, D., & Zhang, B. (2022). Cellxgene VIP unleashes full power of interactive visualization and integrative analysis of scRNA-seq, spatial transcriptomics, and multiome data. bioRxiv. https://doi.org/10.1101/2020.08.28.270652