Blog Categories
RNA-Seq Case Study: iPSC Differentiation into Muscle and Neuron
|
We use BxGenomics to examine RNA-Seq data from three key time points during human iPSC differentiation into mature muscle fibers with motor neurons. Reference: Cells. 2020 Jun 23;9(6):1531. doi: 10.3390/cells9061531. Study GEO ID: GSE142042 |
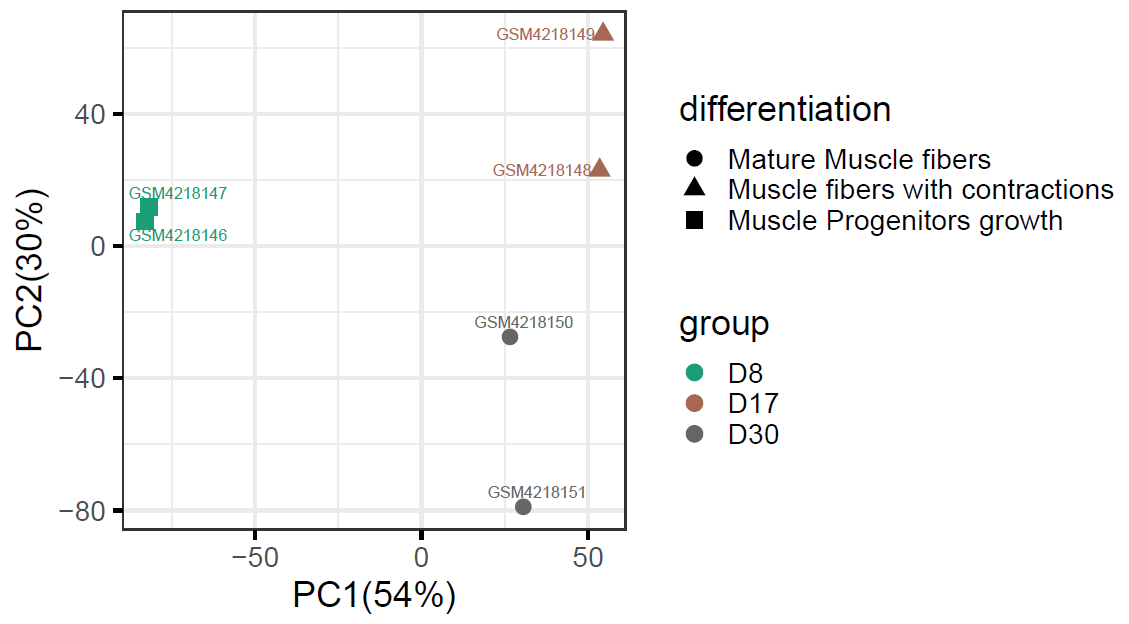 |
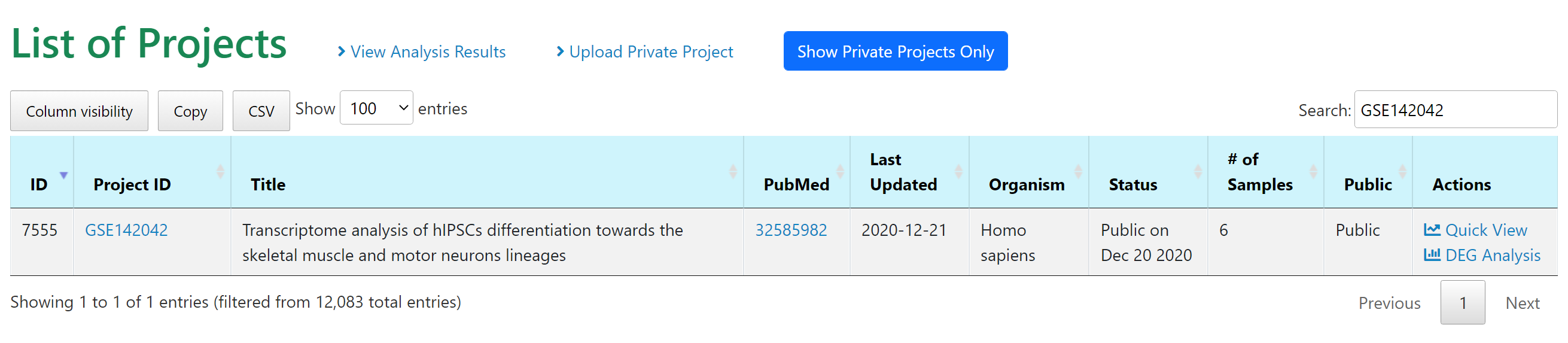
First we use the study GEO ID GSE142042 to find the study, then we can choose "DEG Analysis" under Actions column to perform comparisons between the three time points, D8 (muscle progenitors), D17 (muscle fibers with contractions), and D30 (mature muscle fibers).
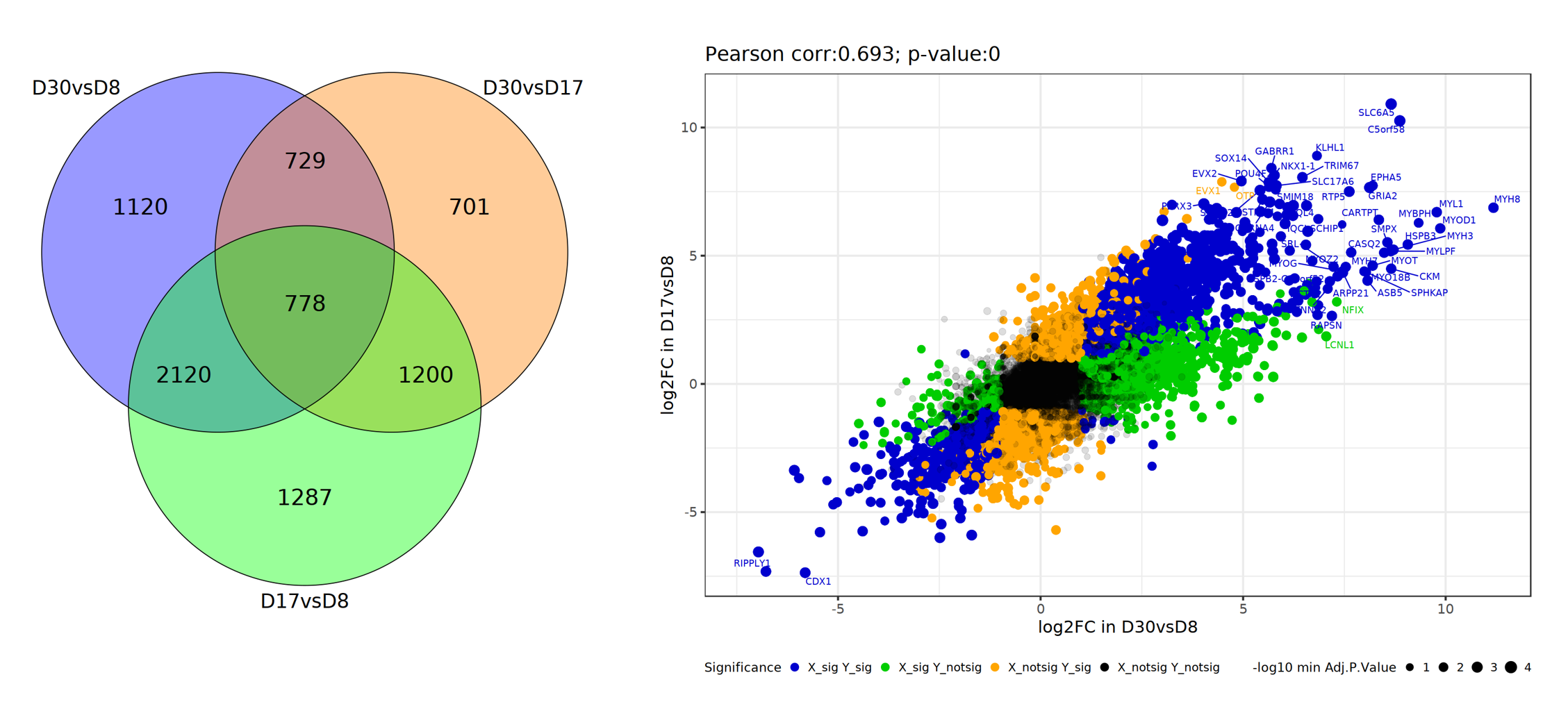
From the analysis results, we can use the Venn Diagram tool to compare the three DEG sets. There are a lot of similarities between D17vsD8 and D30vsD8, and we can use DEGs in two comparisons to further examine these two comparisons.
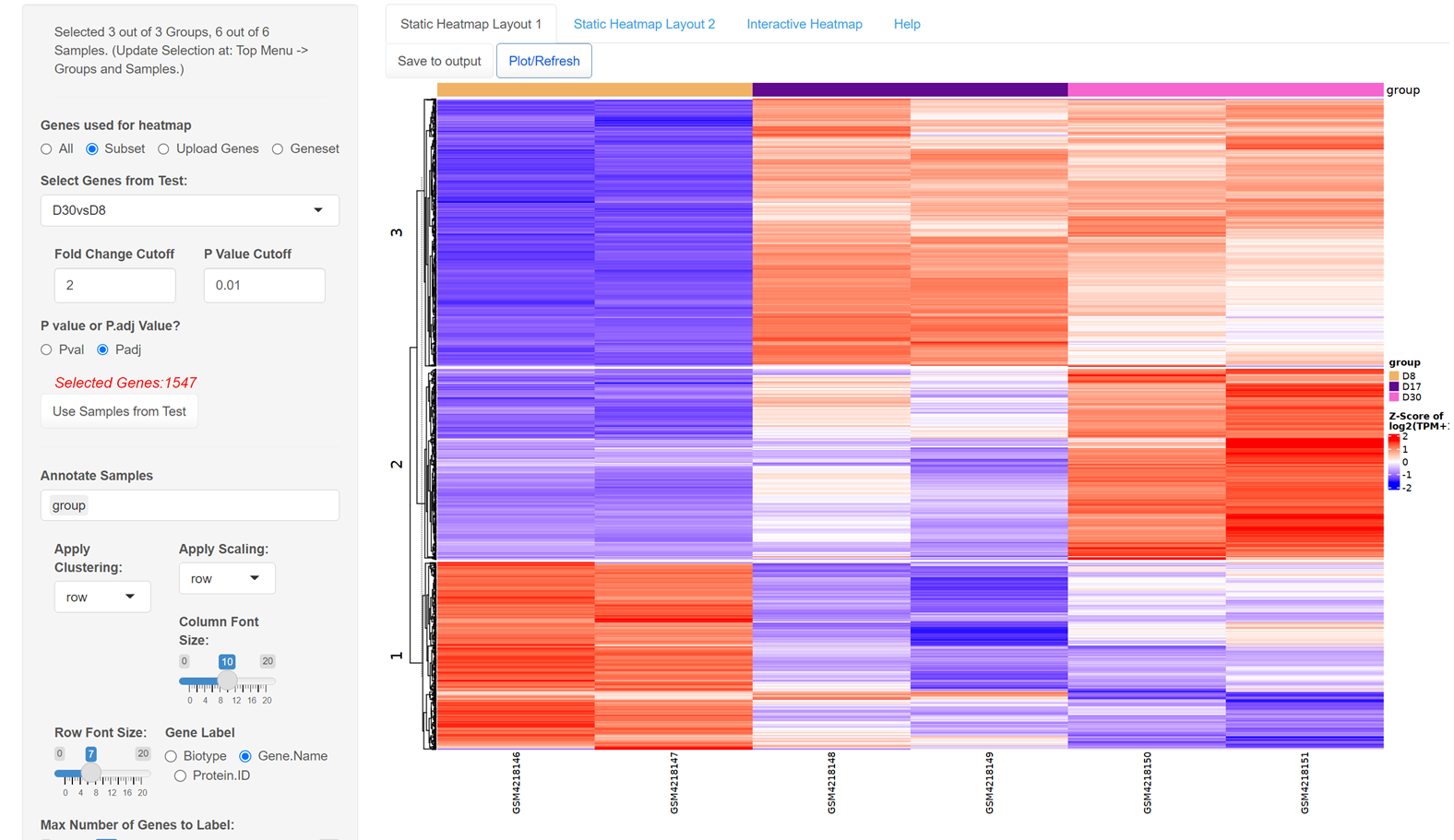
If we plot a heatmap using DEGs from D30vsD8, we can see the DEGs can be roughly divided into three groups, the cut row feature in heatmap options allows us to identify these three groups using k-mean clustering.
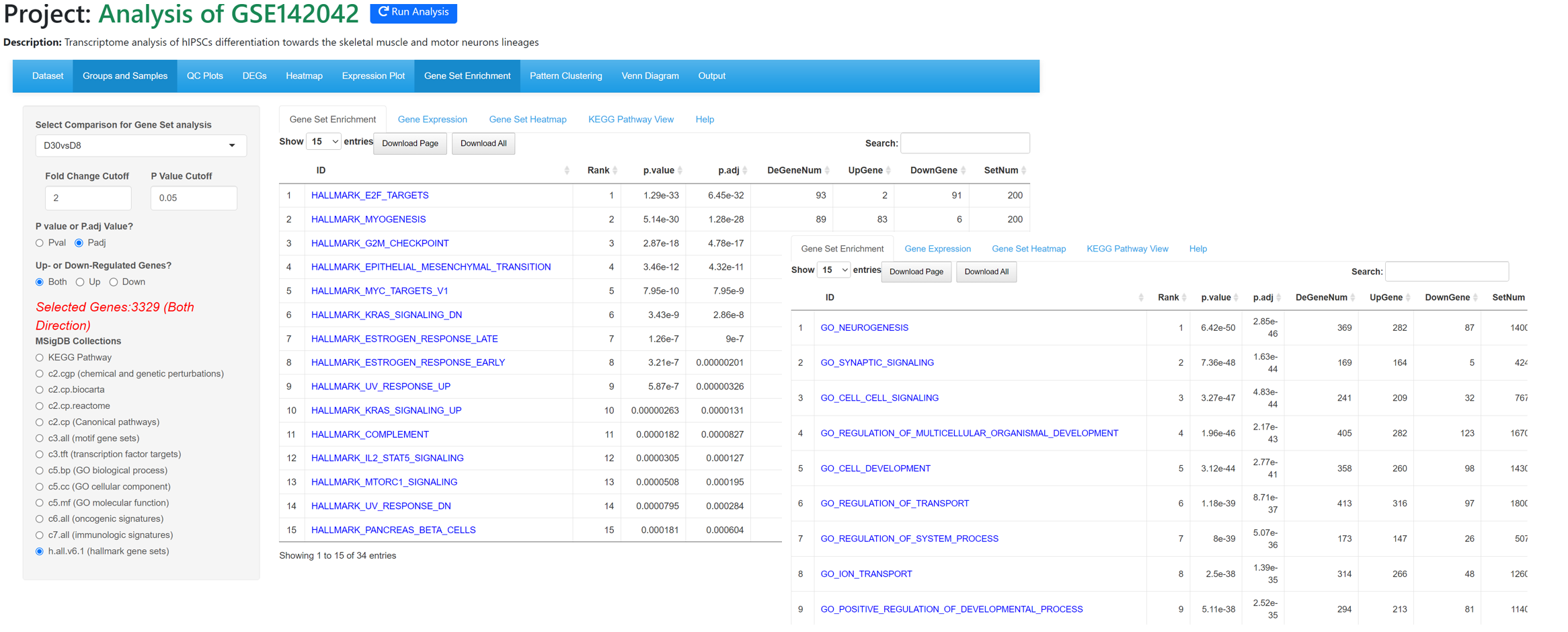
From Gene Set Enrichment tool, we can see myogenesis and nurogenesis genes are highly enriched in D30vsD8 comparison, confirming that the cells have been differentiated into muscle and neuron cells.
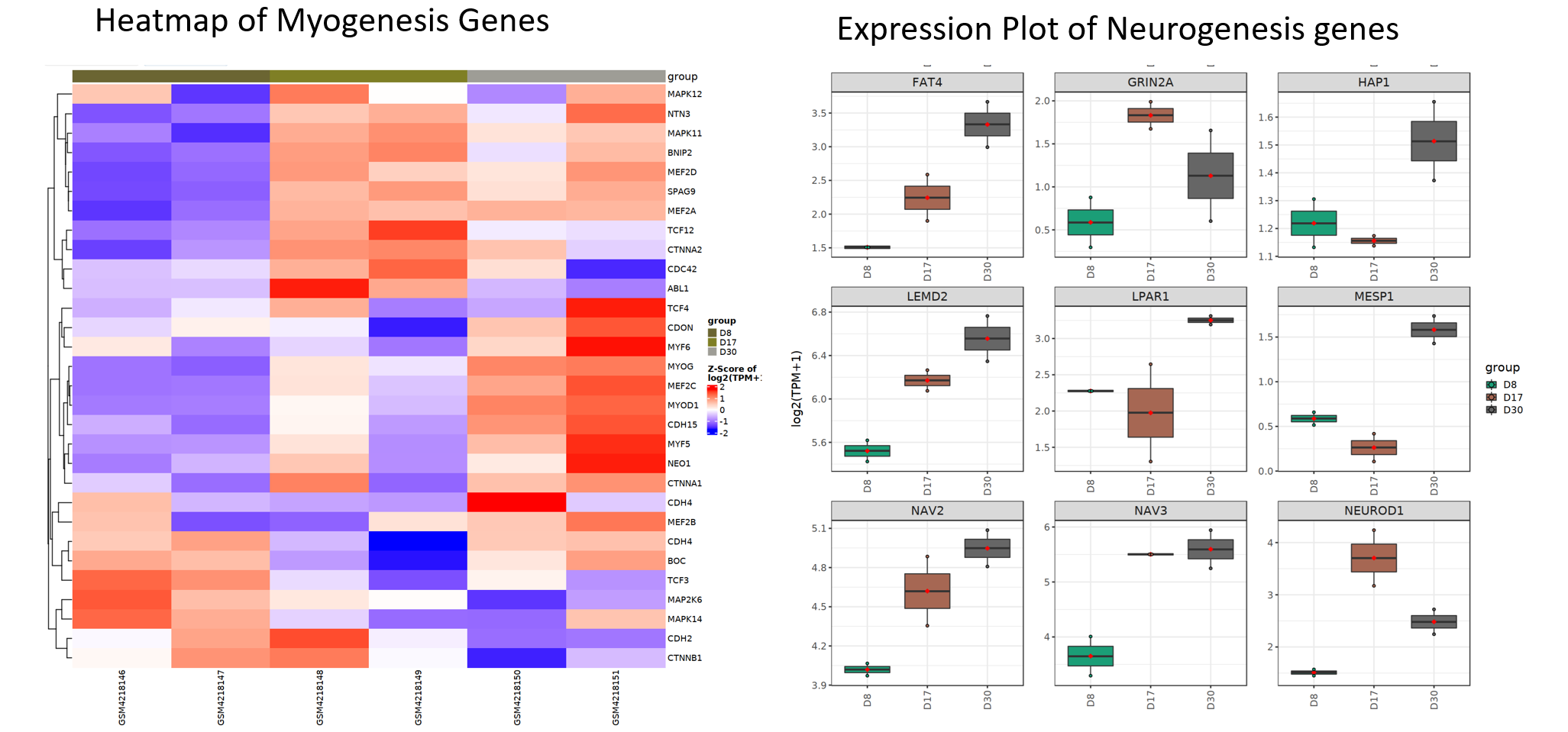
Lastly, we can use plot myogenesis and nurogenesis genes using heatmap or expression plot to view these genes in details.