Genomics: BioInfoRx offers supported solution for a published ecosystem for single-cell RNA data
If you’re interested in using scRNASequest but lack the IT infrastructure or bioinformatics expertise, try the fully supported BxGenomics scRNA-Seq View which is ready to use and requires no installation.
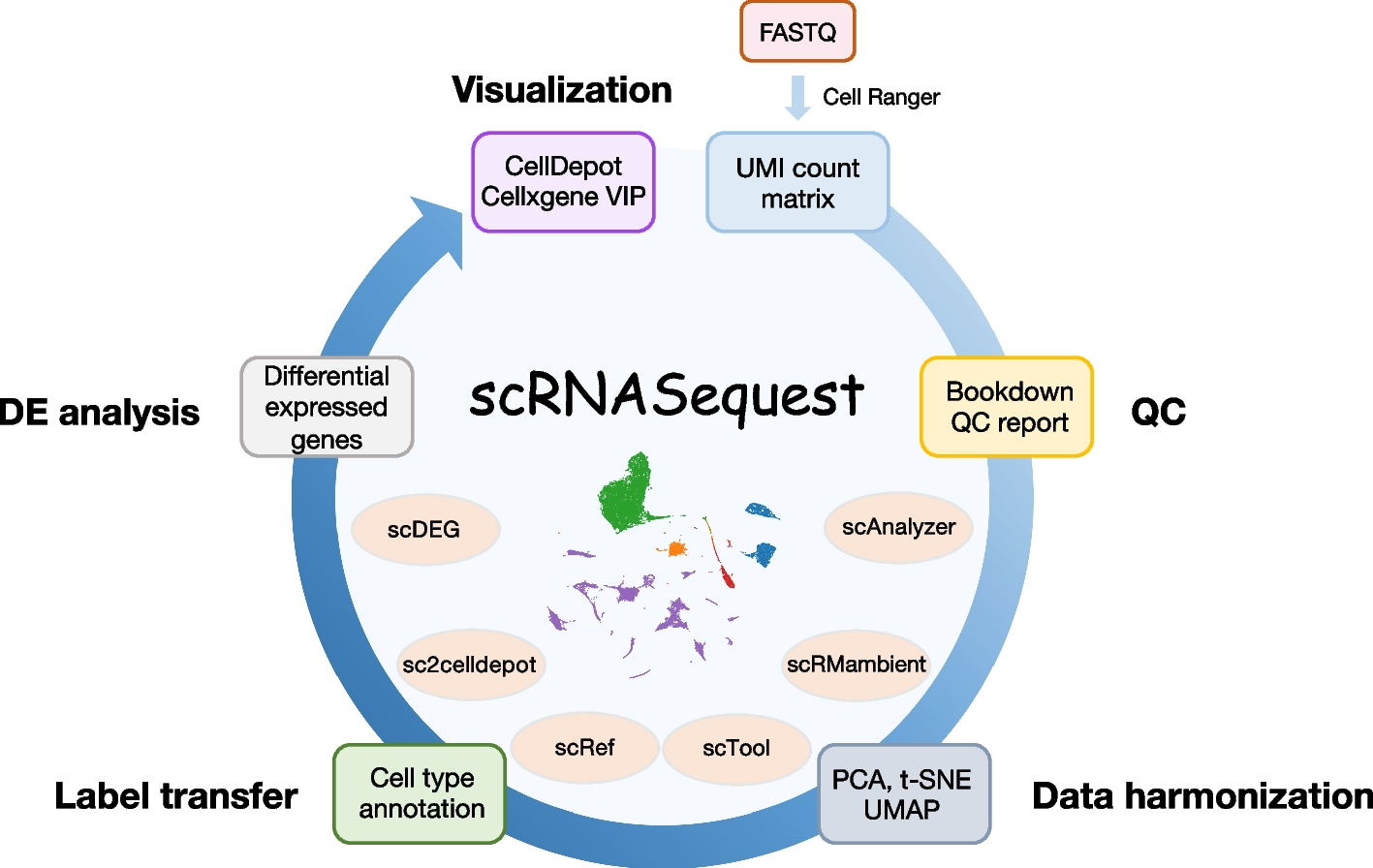
The BioInfoRx team in collaboration with Biogen have recently published a one-stop solution for scRNA-seq data analysis in BMC Genomics, scRNASequest: an ecosystem of scRNA-seq analysis, visualization, and publishing. The scRNASequest pipeline integrates multiple tools to preprocess, harmonize, annotate, compare, and explore scRNA-seq data from different sources and conditions. It generates h5ad files that can be published to online database CellDepot and visualized with cellxgene VIP. scRNASequest is available as an open-source package at GitHub, along with a detailed tutorial.
Recognizing the needs from many researchers who want to analyze their single-cell data with scRNASequest, but lack the IT infrastructure of computational expertise, BioInfoRx is glad to offer commercial support for scRNASequest, as well as additional tertiary analysis to help researchers gain insights from single-cell data. The commercially supported BxGenomics scRNA-Seq system is built upon the latest cloud technology and is optimized to use a secure system to efficiently handle many single-cell data sets from different users. Researchers can try the supported solution at the BxGenomics website at no cost. All accounts can access the over 400 public single-cell data sets for free. Users can upgrade to pro account to have their own single-cell RNA-Seq data processed and uploaded into the system, and take advantage of the many data mining tools that come with the system.
The main difference between the open-source solution and supported solutions are listed in the table below.
| Step | Open-Source Solution (scRNASequest) | Supported Solution (BxGenomics scRNA-Seq View) |
|---|---|---|
| Hardware | Need powerful local or cloud Linux server. | Nothing required for the user. Ready to use. |
| software | Need to install pipeline from GitHub or Docker, need to install and configure web App. | Nothing required for the user. Ready to use. |
| Data Analysis | QC, harmonize, annotate, compare, output h5ad. | All steps in open-source, plus advanced project-specific analysis. |
| Online database | Public database, or users can install local private copy. | Unified database for all public data, plus user’s private data sets under access control. |
| Data Visualization | Users need to install cellxgene VIP, and manually load and unload data sets. | BxGenomics launches cellxgene VIP visualization automatically. Nothing required for the user. |