BxSeqTools Ultimate Molecular Cloning Guides - TA TOPO Cloning
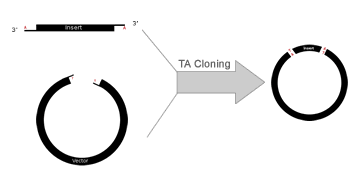
| TA Cloning is a subcloning technique that doesn't the use of restriction enzymes and is easier and quicker than traditional subcloning. The technique relies on the ability of adenine (A) and thymine (T) (complementary basepairs) on different DNA fragments to hybridize and, in the presence of ligase, become ligated together. PCR products are usually amplified using Taq DNA polymerase which preferentially adds an adenine to the 3' end of the product. Such PCR amplified inserts are cloned into linearized vectors that have complementary 3' thymine overhangs. | |||||||||
 |
|||||||||
TA Topo Cloning Construct Design with BxSeqToolsAdvantages of using BxSeqTools for TA Topo cloning design:
|
|||||||||
Overall TA Topo Cloning ProceduresCreating The InsertThe insert is created by PCR using Taq DNA polymerase. This polymerase lacks 5' to 3' proofreading activity and with a high probability adds a single, 3'-adenine overhang to each end of the PCR product. It is best if the PCR primers have guanines at the 5' end as this maximizes probability of Taq DNA polymerase adding the terminal adenosine overhang. Thermostable polymerases containing extensive 3´ to 5´ exonuclease activity should not be used as they do not leave the 3´ adenine-overhangs. Creating the VectorThe target vector is linearized and cut with a blunt-end restriction enzyme. This vector is then tailed with dideoxythymidine triphosphate (ddTTP) using terminal transferase. It is important to use ddTTP to ensure the addition of only one T residue. This tailing leaves the vector with a single 3'-overhanging thymine residue on each blunt end. Manufacturers commonly sell TA Cloning "kits" with with a wide range of prepared vectors that have already been linearized and tagged with an overhanging thymine residue.
TA-TOPO Cloning is a cloning method that takes advantage of the terminal transferase activity of some DNA Polymerases that add a 3’-dA overhang to each end of a PCR product. A linearized vector with prepared 3’-T overhangs facilitates ligation of the PCR product.
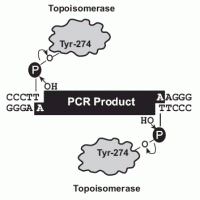
Technical Notes: Taq polymerase has a nontemplate-dependent terminal transferase activity that adds a single deoxyadenosine (A) to the 3´ ends of PCR products. The linearized vector supplied has single, overhanging 3´ deoxythymidine (T) residues. This allows PCR inserts to ligate efficiently with the vector. Topoisomerase I from Vaccinia virus binds to duplex DNA at specific sites and cleaves the phosphodiester backbone after 5'-CCCTT in one strand. The energy from the broken phosphodiester backbone is conserved by formation of a covalent bond between the 3' phosphate of the cleaved strand and a tyrosyl residue (Tyr-274) of topoisomerase I. The phospho-tyrosyl bond between the DNA and enzyme can subsequently be attacked by the 5' hydroxyl of the original cleaved strand, reversing the reaction and releasing topoisomerase.
|
|||||||||
| Read more on Wikipedia about TA TOPO Cloning | |||||||||
References
|
|||||||||