BxSeqTools Ultimate Molecular Cloning Guides - Gene Synthesis Cloning
| Gene Synthesis Cloning is a technique used to move a particular gene of interest from a parent vector to a destination vector in order to further study its functionality. |
 |
 |
Gene Synthesis Cloning construct design with BxSeqToolsAdvantages of using BxSeqTools for gene synthesis design:
|
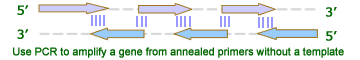
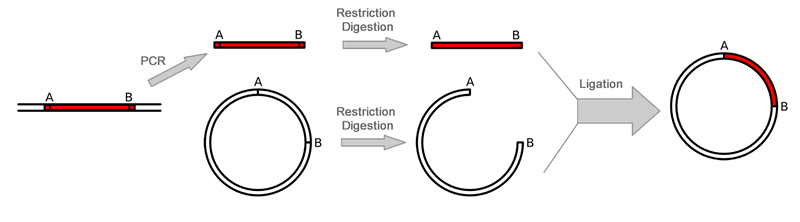
Overall Gene Synthesis Cloning ProceduresGene synthesis cloning is similar to PCR Cloning except that the gene is synthesized through PCR reaction with overlapped primers. There is no template is needed. This is useful when only protein sequence is known or users like to optimize codons. The gene is first amplified using Polymerase Chain Reaction (PCR) after mixing the overlaping primers. Restriction enzymes are then used to excise the gene of interest (the insert) from the parent. The insert is purified in order to isolate it from background junk. A common purification method is gel isolation. Simultaneously, the same or compatible restriction enzymes are used to digest (cut) the destination. The idea behind using the same restriction enzymes is to create complementary sticky ends, which will facilitate ligation later on. A phosphatase (commonly Calf Intestinal Alkaline Phosphatase; CIAP) is also added to prevent self-ligation of the destination vector. The digested destination vector is isolated/purified. The insert and the destination vector are then mixed together with DNA ligase. A typical ratio of insert genes to destination vectors is 3:1; by increasing the insert concentration, self-ligation is further decreased. After letting the reaction mixture sit for a set amount of time at a specific temperature (dependent upon the size of the strands being ligated; for more information see DNA ligase), the insert should become successfully incorporated into the parent plasmid.
|