BxSeqTools Ultimate Molecular Cloning Guides - Directional TOPO Cloning
| Use PCR reaction to amplify a DNA fragment. The resulting PCR products have four additional bases (CACC) at the 5' ends that are from the specially designed forward PCR primer. With a special ligation kit, this fragment is directly ligated into a linearized vector DNA (D-TOPO Vector, which contains GTGG overhangs at the 5' end) without pre-digestion with restriction endonucleases. The fragment can only be inserted in forward orientation. | ||||||||
 |
||||||||
Directional TOPO Cloning construct design with BxSeqToolsAdvantages of using BxSeqTools for subcloning design:
|
||||||||
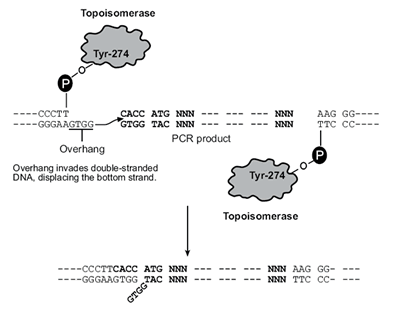
Overall Directional TOPO Cloning ProceduresDirectional TOPO® Cloning is a method that utilizes the binding activity of Topoisomerase I. A sequence of interest is PCR amplified using a forward strand primer with four additional bases (CACC). Commercial vector sequences have been engineered with topoisomerase bound at 3’ and a complementary 5’ GTGG overhang. The overhang invades the 5’ end of the PCR product and anneals to the added bases. The 5’ hydroxyl group of the PCR product attacks the topoisomerase bound to the vector, releasing it and causing the ligation of the phosphate backbone. Orientation of the insert is determined by the 5’ overhang, allowing the other end to be blunt.
Technical Notes: In this system, PCR products are directionally cloned by adding four bases to the forward primer (CACC). The overhang in the cloning vector (GTGG) invades the 5′ end of the PCR product, anneals to the added bases, and stabilizes the PCR product in the correct orientation. Directional joining of double-strand DNA using TOPO-charged oligonucleotides occurs by adding a 3′ single-stranded end (overhang) to the incoming DNA. This single-stranded overhang is identical to the 5′ end of the TOPO-charged DNA fragment. At Invitrogen, this idea has been modified by adding a 4 nucleotide overhang sequence to the TOPO-charged DNA and adapting it to a ‘whole vector’ format.
|
||||||||
|